安装
1 | pip install -U scikit-learn |
安装 graphviz 一个对dot文件进行绘图的软件,可以用于对模型进行可视化。
数据准备
数据准备包括训练集和测试集两部分的数据准备(也可以是一批数据按比例进行拆分,一部分作为训练一部分作为测试)
这里可以下载一份测试数据
数据格式如下(前30行):1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32id,N_Days,Drug,Age,Sex,Ascites,Hepatomegaly,Spiders,Edema,Bilirubin,Cholesterol,Albumin,Copper,Alk_Phos,SGOT,Tryglicerides,Platelets,Prothrombin,Stage,Status
0,999,D-penicillamine,21532,M,N,N,N,N,2.3,316.0,3.35,172.0,1601.0,179.8,63.0,394.0,9.7,3.0,D
1,2574,Placebo,19237,F,N,N,N,N,0.9,364.0,3.54,63.0,1440.0,134.85,88.0,361.0,11.0,3.0,C
2,3428,Placebo,13727,F,N,Y,Y,Y,3.3,299.0,3.55,131.0,1029.0,119.35,50.0,199.0,11.7,4.0,D
3,2576,Placebo,18460,F,N,N,N,N,0.6,256.0,3.5,58.0,1653.0,71.3,96.0,269.0,10.7,3.0,C
4,788,Placebo,16658,F,N,Y,N,N,1.1,346.0,3.65,63.0,1181.0,125.55,96.0,298.0,10.6,4.0,C
5,703,D-penicillamine,19270,F,N,Y,N,N,0.6,227.0,3.46,34.0,6456.2,60.63,68.0,213.0,11.5,3.0,D
6,1300,Placebo,17703,F,N,N,N,N,1.0,328.0,3.35,43.0,1677.0,137.95,90.0,291.0,9.8,3.0,C
7,1615,Placebo,21281,F,N,Y,N,N,0.6,273.0,3.94,36.0,598.0,52.7,214.0,227.0,9.9,3.0,C
8,2050,D-penicillamine,20684,F,N,N,N,N,0.7,360.0,3.65,72.0,3196.0,94.55,154.0,269.0,9.8,2.0,C
9,2615,D-penicillamine,15009,F,N,N,N,N,0.9,478.0,3.6,39.0,1758.0,171.0,140.0,234.0,10.6,2.0,C
10,3581,Placebo,25772,F,N,N,N,N,0.5,252.0,3.6,26.0,377.0,56.76,185.0,336.0,10.0,2.0,C
11,1614,Placebo,14106,F,N,N,N,N,0.9,328.0,3.61,62.0,1105.0,137.95,95.0,145.0,9.5,3.0,C
12,1847,Placebo,12279,F,N,N,N,N,0.6,232.0,3.68,38.0,1029.0,128.65,99.0,273.0,10.7,2.0,C
13,1153,D-penicillamine,22347,F,N,Y,N,N,0.6,232.0,3.83,24.0,678.0,65.1,99.0,248.0,10.4,3.0,C
14,904,D-penicillamine,22388,F,N,Y,N,N,3.9,304.0,3.2,13.0,1440.0,153.45,169.0,156.0,10.0,3.0,D
15,1212,Placebo,15112,F,N,N,N,N,0.7,335.0,3.54,44.0,1345.0,137.95,145.0,244.0,10.6,3.0,C
16,1967,Placebo,17884,F,N,N,N,N,0.7,328.0,3.58,39.0,1065.0,98.0,78.0,259.0,11.7,2.0,C
17,1592,D-penicillamine,14872,F,N,Y,N,N,1.1,392.0,3.43,39.0,1395.0,184.45,133.0,328.0,11.2,2.0,C
18,1481,Placebo,18302,F,N,N,N,N,1.0,259.0,3.85,67.0,936.0,134.85,139.0,341.0,9.6,3.0,C
19,3358,Placebo,17031,F,N,N,N,N,0.6,322.0,3.77,52.0,834.0,60.45,214.0,153.0,11.0,3.0,C
20,3092,Placebo,15612,F,N,Y,N,N,0.6,303.0,3.1,70.0,1032.0,56.76,154.0,336.0,10.6,4.0,C
21,326,D-penicillamine,18199,F,N,Y,Y,S,6.6,244.0,3.02,199.0,1819.0,170.5,91.0,132.0,12.1,4.0,D
22,2363,Placebo,17703,F,N,N,N,N,1.0,215.0,3.95,58.0,645.0,97.65,71.0,233.0,10.1,4.0,C
23,1152,D-penicillamine,16736,F,N,Y,N,N,1.1,373.0,3.9,69.0,1353.0,116.25,139.0,268.0,10.0,4.0,C
24,3577,D-penicillamine,27398,F,N,N,N,N,0.6,253.0,4.03,38.0,642.0,41.85,112.0,227.0,9.9,2.0,C
25,799,Placebo,27220,F,N,Y,N,N,1.3,325.0,3.6,81.0,2065.0,232.5,100.0,277.0,11.1,4.0,C
26,1832,Placebo,17442,F,N,Y,N,N,2.0,328.0,3.35,76.0,2276.0,114.7,104.0,518.0,10.0,4.0,D
27,4467,D-penicillamine,12398,F,N,Y,N,N,1.2,414.0,3.43,41.0,876.0,84.0,110.0,385.0,11.0,3.0,C
28,2301,D-penicillamine,15105,F,N,Y,Y,N,2.3,528.0,3.34,173.0,1282.0,120.9,55.0,123.0,10.7,4.0,D
29,943,Placebo,19002,F,N,N,N,N,28.0,556.0,3.26,39.0,1713.0,171.0,171.0,348.0,10.2,3.0,D
30,1882,Placebo,15265,F,N,Y,Y,N,1.1,316.0,3.35,67.0,1353.0,137.95,137.0,273.0,9.6,3.0,C
打眼一看,我们可以看到数据中有很多文本/类别的信息是使用字符串进行表示的,所以在进行文本读取后,我们需要对数据进行一个整体的预处理过程。1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28import pandas as pd
trains_data = pd.read_csv("train.csv",sep=",")
test_data = pd.read_csv("test.csv",sep=",")
# 删除一些不会考虑作为训练指标的信息,例如Status是最终结果,Edema指标是分散文本。
Trains_input= trains_data.drop(["Status","Edema"],axis=1)
Trains_target= trains_data["Status"]
# 对其他一些分类指标构建数据清洗映射关系
Drug_mapping = {"D-penicillamine":1,"Placebo":2}
sex_mapping = {'F': -1, 'M': 1}
Ascites_mapping = {"N":0,"Y":1}
Hepatomegaly_mapping = {"N":0,"Y":1}
Spiders_mapping = {"N":0,"Y":1}
# 对文本信息进行更新迭代
Trains_input["Drug"]=Trains_input["Drug"].replace(Drug_mapping)
Trains_input["Sex"]=Trains_input["Sex"].replace(sex_mapping)
Trains_input["Ascites"]=Trains_input["Ascites"].replace(Ascites_mapping)
Trains_input["Hepatomegaly"]=Trains_input["Hepatomegaly"].replace(Hepatomegaly_mapping)
Trains_input["Spiders"]=Trains_input["Spiders"].replace(Spiders_mapping)
Trains_input.head()
# 结果示例
id N_Days Drug Age Sex Ascites Hepatomegaly Spiders Bilirubin Cholesterol Albumin Copper Alk_Phos SGOT Tryglicerides Platelets Prothrombin Stage
0 0 999 1 21532 1 0 0 0 2.3 316.0 3.35 172.0 1601.0 179.80 63.0 394.0 9.7 3.0
1 1 2574 2 19237 -1 0 0 0 0.9 364.0 3.54 63.0 1440.0 134.85 88.0 361.0 11.0 3.0
2 2 3428 2 13727 -1 0 1 1 3.3 299.0 3.55 131.0 1029.0 119.35 50.0 199.0 11.7 4.0
数据概览
1 | import sweetviz |
模型训练
1 | # 模型(也可用单个决策树) |
基于此,我们就完成了一个最基本的模型构建,也可以通过这个模型进行简单的数据分析/预测,完成了入门,当然这个随便确定参数的模型性能可能并不完善,所以接下来我们就要看看怎么进行调参优化。
指标筛选
指标权重
1 | from sklearn.ensemble import RandomForestClassifier |
就可以得到每个指标在最终建模时的权重(权重综合为1),格式如下1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17 1) Bilirubin 0.195250
2) N_Days 0.149384
3) Copper 0.091069
4) Prothrombin 0.085256
5) Age 0.064164
6) SGOT 0.063270
7) Platelets 0.057314
8) Cholesterol 0.054671
9) Albumin 0.052882
10) Alk_Phos 0.052205
11) Tryglicerides 0.048500
12) Stage 0.031570
13) Hepatomegaly 0.020866
14) Spiders 0.013486
15) Drug 0.009804
16) Sex 0.006664
17) Ascites 0.003645
当变量参数非常多的时候,我们可以基于每个指标的权重进行筛选,对输入参数进行精简。比如在我们这个数据集中,Bilirubin,N_Days等指标重要性比较高,而对应的Ascites、Sex、Drug等指标重要性就相对较低。
参数调优
逐步调优
借助交叉验证得分确定最有决策树数目。
在随机森林里面,决策树越多,在对训练集的建模表现越好,但是对应的资源需求也就越大。所以有时候,我们需要进行评估,构建多少颗决策树更合适。这时候,我们可以使用 cross_val_score 进行不同梯度交叉验证1
2
3
4
5
6
7
8
9
10
11
12superpa=[]
#for i in [1,10,20,30,40,50,60,70,80,90,100,110,120,130,140,150,160,170,180,190,200]:
for i in range(20):
#rfc = RandomForestClassifier(max_depth=i+1,n_jobs=-1,random_state=1,n_estimators=i+1)
rfc = RandomForestClassifier(n_estimators=i+1,random_state=1)
rfc_s = cross_val_score(rfc,Trains_input,Trains_target,cv=10).mean()
superpa.append(rfc_s)
# 打印分数最高时的分数和max_depth的值
print(max(superpa),superpa.index(max(superpa))+1)
plt.figure(figsize=[20,5])
plt.plot(range(1,201),superpa)
plt.show()
确定决策树数目后,确定决策树深度
基于上述确定的决策数数目,对决策树的最大深度进行遍历,确定最大决策树深度1
2
3
4
5
6
7
8
9
10superpa=[]
for i in range(20):
rfc = RandomForestClassifier(max_depth=i+1,n_jobs=-1,random_state=1,n_estimators=70)
rfc_s = cross_val_score(rfc,Trains_input,Trains_target,cv=10).mean()
superpa.append(rfc_s)
# 打印分数最高时的分数和max_depth的值
print(max(superpa),superpa.index(max(superpa))+1)
plt.figure(figsize=[20,5])
plt.plot(range(1,21),superpa)
plt.show()
网格调优
1 | from sklearn.ensemble import RandomForestClassifier |
模型可视化
1 |
|
得到的随机森林某一个决策树判断逻辑如下: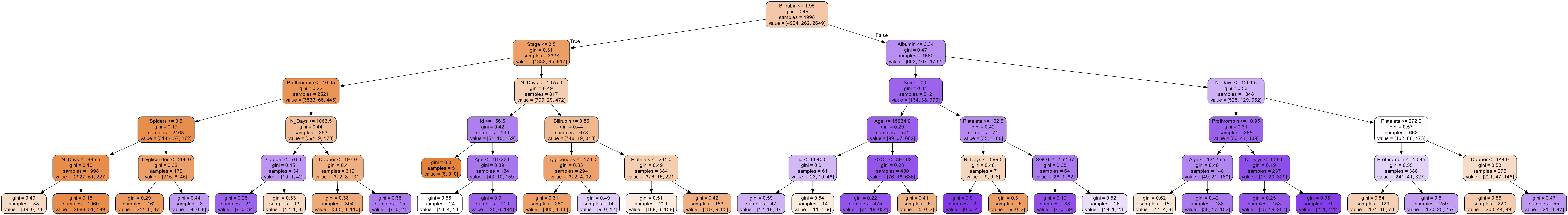
模型应用
通过上述步骤,我们已经成功的得到我们所需要的一个随机森林模型 model ,杰西莱我们就需要使用我们的模型,对于全新的测试数据进行处理。1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21# 因为一般训练集和测试集的数据格式是一样的,所以我们对训练集进行的数据处理过程需要同时对测试集进行处理。
Drug_mapping = {"D-penicillamine":1,"Placebo":2}
sex_mapping = {'F': -1, 'M': 1}
Ascites_mapping = {"N":0,"Y":1}
Hepatomegaly_mapping = {"N":0,"Y":1}
Spiders_mapping = {"N":0,"Y":1}
test_data["Drug"]=test_data["Drug"].replace(Drug_mapping)
test_data["Sex"]=test_data["Sex"].replace(sex_mapping)
test_data["Ascites"]=test_data["Ascites"].replace(Ascites_mapping)
test_data["Hepatomegaly"]=test_data["Hepatomegaly"].replace(Hepatomegaly_mapping)
test_data["Spiders"]=test_data["Spiders"].replace(Spiders_mapping)
test_data= test_data.drop(["Edema","id"],axis=1)
test_data.head()
#直接返回预测结果
model.predict(test_data)
# 返回每类的预测概率
result = model.predict_proba(test_data)
result_df =pd.DataFrame(result,columns = ['Status_C', 'Status_CL', 'Status_D'])