Publish
原文链接: CNVkit Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing-annotated
cnvkit.readthedocs
原理介绍PPT
CNVKit若干算法问题详解
检测原理在上述文献、PPT中已经有相对详细的介绍,此处不再进行展开。针对目前一些可视化的需求,补充记录一些关于CNVKit结果展示的相关功能和使用方法。
可视化
CNVKit提供了比较详细的可视化模块
三种可视化的主体绘图命令如下:1
2
3cnvkit.py scatter -h
cnvkit.py diagram -h
cnvkit.py heatmap -h
scatter
说明
1 | cnvkit.py scatter Sample.cnr -s Sample.cns |
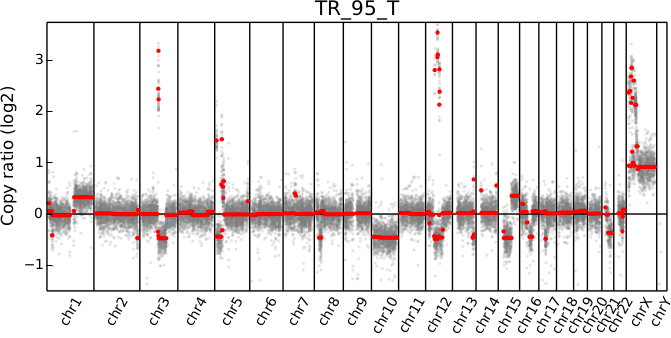
扩展参数1
2
3
4
5
6
7python cnvkit.py scatter Sample.cnr
-s Sample.cns
-c chr7 # 指定参考基因组,可以进一步细化指定具体范围chr5:100-50000000
-g BRAF,MET # 指定展示的基因,多个基因使用逗号(",")分隔
-v Sample.vcf #
```
## 示例
/jdfstj1/B2C_COM_P1/PipeAdmin/02.software/Conda/bin/python /jdfstj1/B2C_COM_P1/PipeAdmin/04.Pipeline/01.AIO.v2.01/bin/cnvkit.py scatter ../Analyze/cnv/pancancer689__DX1790_huangrenhua_20S8320498R_20B8320498__Cancer.markdup.cnr -s ../Analyze/cnv/pancancer689__DX1790_huangrenhua_20S8320498R_20B8320498__Cancer.markdup.cns -c chr3:150000000-220000000 -g EIF4A2,TIPARP,TP631
2
3
4
5
6
7
## diagram
```shell
python cnvkit.py diagram Sample.cnr
-s Sample.cns
-t 2 # 对判定为CNV的阈值进行调整(仅达到阈值的基因会进行单独展示)
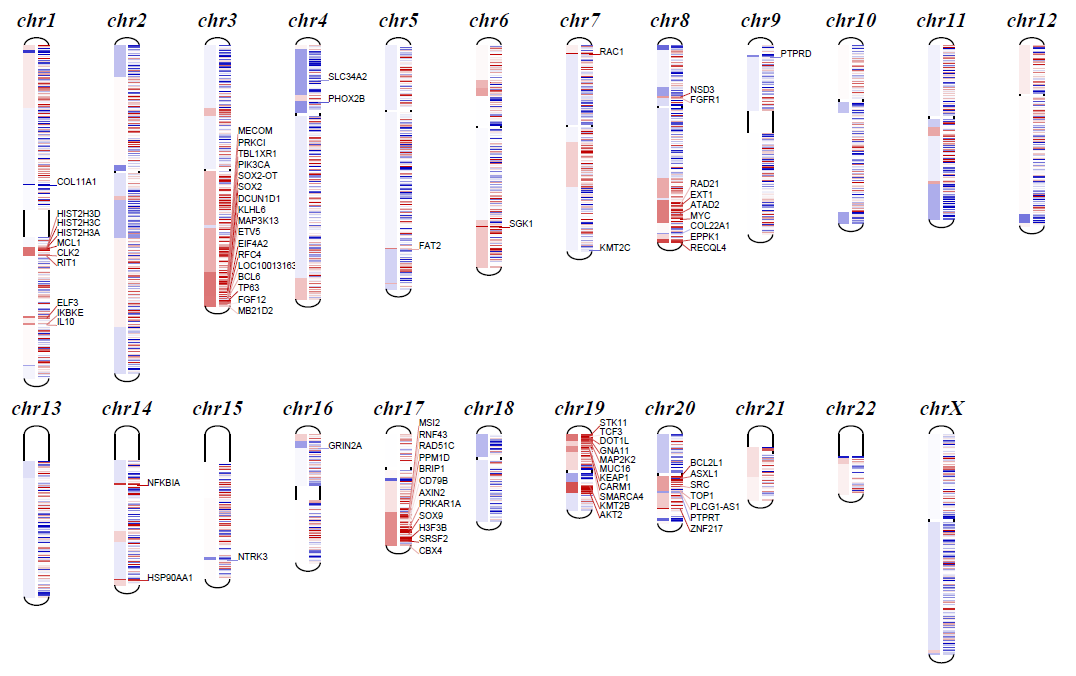
结果格式说明
*.cnv
1 | chromosome, start, end, gene, log2, depth and weight |
异常处理记录
Qt载入失败
具体报错示例如下:
1
2
3
4
5
6
7
8Reinstalling the application may fix this problem.
Showing 918 probes and 1 selected genes in region chr15
This application failed to start because it could not find or load the Qt platform plugin "xcb"
in "".
Available platform plugins are: eglfs, minimal, minimalegl, offscreen, xcb.
Reinstalling the application may fix this problem.处理解决方案