软件文章
DECoN软件发表文献
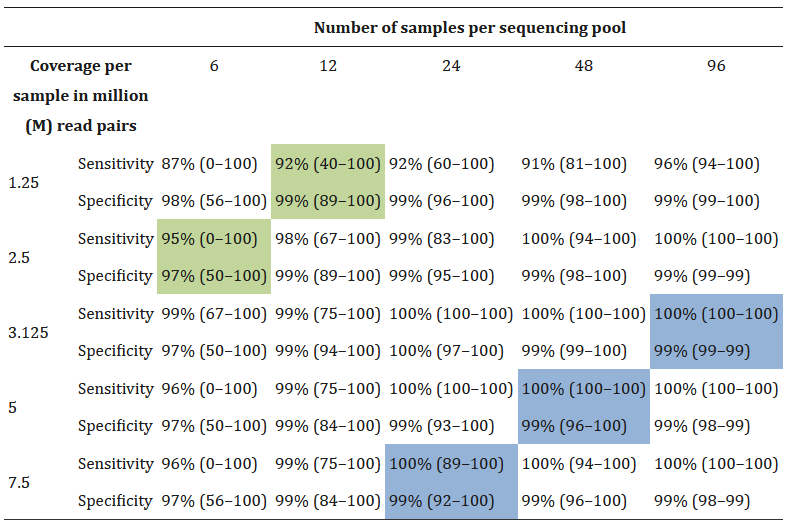
参考文献Accurate clinical detection of exon copy number variants in a targeted NGS panel using DECoN
综述类文章
参考文献:2025 Advocating Targeted Sequential Screening over Whole Exome Sequencing in 21-Hydroxylase Deficiency
参考文献:Evaluation of three read-depth based CNV detection tools using whole-exome sequencing data
cite: Yao, Ruen et al. “Evaluation of three read-depth based CNV detection tools using whole-exome sequencing data.” Molecular cytogenetics vol. 10 30. 23 Aug. 2017, doi:10.1186/s13039-017-0333-5
CoNIFER and XHMM have an even poorer detection sensitivity for smaller CNVs involving fewer capturing probes, whereas CNVnator had a rather consistent detection sensitivity for all size CNVs(> 3 kb).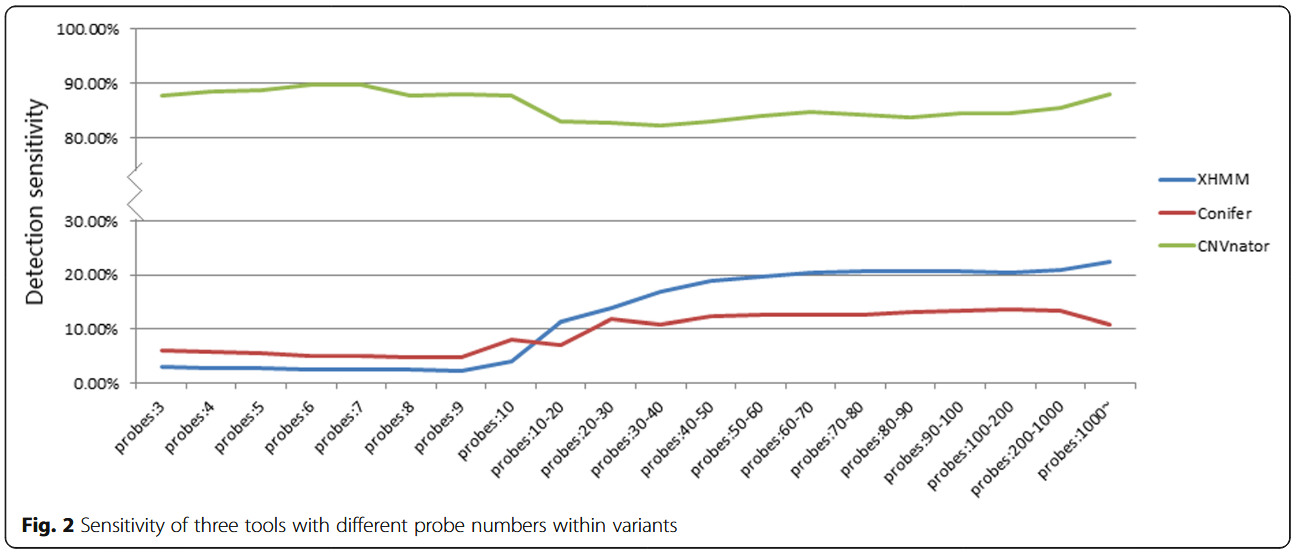
总结: 灵敏性整体不到90%。(涵盖大片段CNV)
参考文献:Identification of Copy Number Variants Through Whole-Exome Sequencing in Autosomal Recessive Nonsyndromic Hearing Loss
CoNIFER 检测到的杂合假阳性缺失(40%)(区域大小为 3E1 kb)比例低于 XHMM(64%)
参考文献: Exome sequence read depth methods for identifying copy number changes
提供的详细原表
Paired data 配对数据
| Program 程序(参考) | 性能数据 |
| ——————————————– | ———————————————————————————————————————————————— |
| ExomeCNV | 对于 WGS 数据,缺失的敏感性分别为 86% ;重复的敏感性为 88% |
| exome2cnv | 灵敏度超过 89%,假阳性率低于 ExomeCNV |
| PropSeg (proportionality based segmentation) | 与 ExomeCNV 相比,精确度提高了 13%,灵敏度没有差异。 |
| CoNIFER | 检出了 86% 的已验证 CNV,而 ExomeCNV 仅识别出了 14% |
| ExomeDepth | 与 exomeCopy 和 ExomeCNV 相比,检测到的已知 CNV 百分比更高(分别为 75.2%、52.8% 和 41.2%) |
| XHMM | 灵敏度为 67%–92%,较长的 CNV 灵敏度更高。与 CoNIFER 相比,孟德尔遗传定律违背率更低,假阳性率也更少。 |
| Excavator | 与 XHMM 和 CoNIFER 相比,灵敏度更高(分别为 45–55% 和 <20% 和 <5%)。与 XHMM 和 Exome CNV 相比,精确度更高(分别为 30–45% 和 <30% 和 <5%)。 |
| CONTRA | 与 ExomeCNV 相比,外显子敏感性更高(96.4% 对 62.2%) |
| ADTEx (Aberration Detection in Tumour Exome) | 与 VarScan、exomeCopy 和 CONTRA 相比,TP 的检测率更高(分别为 90%、86%、0% 和 0%)。 |
| FishingCNV | 在检测外显子水平的纯合缺失和杂合重复方面,该方法比 CoNIFER 更灵敏(分别为 93% vs 8%和 75% vs 0%)。 |